Antibiotic resistance genes are spread more widely between bacteria than previously thought
A new study published in The Lancet Microbe has found that the transfer of antibiotic resistance genes between different bacteria is considerably more widespread than previously thought.
Researchers at the Ineos Oxford Institute for antimicrobial research (IOI) at Oxford University and Fujian Agriculture and Forestry University in China have developed a new approach, called culture-independent conjugation, to study the transmission of plasmids between bacteria sampled from hospital wastewater.
Plasmids are small circular pieces of DNA that move between neighbouring bacteria (the ‘donor’ bacteria giving the plasmid and the ‘recipient’ bacteria receiving the plasmid) in a process called conjugation. Plasmids containing antibiotic resistance genes are a major driver of antimicrobial resistance (AMR), including resistance against carbapenem- a ‘last-resort’ antibiotic. Carbapenem-resistant bacteria are thought to cause between 50 000 and 100 000 deaths worldwide each year.
This research developed a novel method to further our understanding on the spread of AMR - where and how plasmids are shared between bacteria, particularly plasmids carrying clinically important antibiotic resistance genes. Future work will further examine environmental stressors including those associated with climate change and environmental pollution.
Professor Timothy Walsh, Director of Biology at the Ineos Oxford Institute for antimicrobial research
Up until now, conjugation has been mainly measured with pure cultures containing only one species of bacteria. This has limited understanding of how AMR spreads in bacterial communities in humans, animals, and the environment.
This novel technique, which took over five years to develop, enables conjugation to be assessed in communities of mixed bacteria species, simulating environmental conditions. It is also the first of its kind to model the spread of multidrug resistant plasmids using non-culture methods. The technique can also be readily applied to any aquatic or semi-aquatic samples. Plasmids are tagged with a gene encoding a fluorescent protein; recipient bacteria express the gene and can then be separated using a fluorescence-based sorting method.
The researchers investigated a plasmid called IncX3 (pX3_NDM-5) which carries carbapenem-resistance genes (called NDM-5). This has become globally dominant; however, until now, its spread between bacteria was poorly understood. The researchers used culture-independent conjugation to investigate the transmission of pX3_NDM-5 plasmids between bacteria sampled from wastewater from a hospital in Fuzhou, China.
The study found that this plasmid and others had a much broader host-range and was spread across many types of bacteria including completely unrelated recipient bacteria. This included bacteria which are difficult to culture, suggesting such microbes could be potential plasmid reservoirs contributing to the spread of AMR.
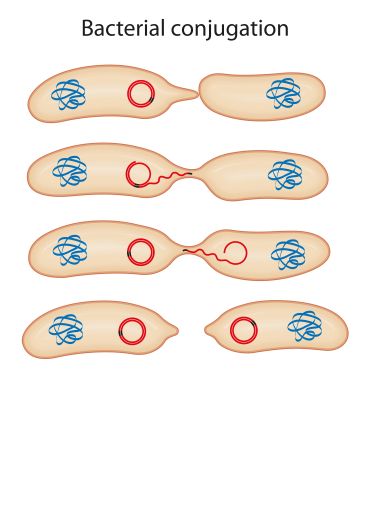 Diagram showing how plasmids are transferred between bacteria (conjugation). Genetic material is transferred between neighbouring bacterial cells by direct cell-to-cell contact or by a bridge-like connection. Image credit: Aldona, Getty Images.
Diagram showing how plasmids are transferred between bacteria (conjugation). Genetic material is transferred between neighbouring bacterial cells by direct cell-to-cell contact or by a bridge-like connection. Image credit: Aldona, Getty Images.These results also break the common assumption that bacteria which are extremely resistant to antibiotics, Gram-negative bacteria, cannot readily exchange antibiotic resistance genes with Gram-positive bacteria – the latter being easier to treat with antibiotics.
The study also explored the effect of environmental stressors on plasmid transfer, finding that chlorine can modify plasmid conjugation. Chlorine is commonly used as a hospital disinfectant and enters hospital wastewater systems, indicating a need to consider the usage of such disinfectants in the face of increasing AMR.
Co-author Professor Timothy Walsh, Director of Biology at the IOI said: ‘We know that overuse and misuse of antibiotics in humans contributes to antimicrobial resistance. This study proves that AMR is being spread in the environment around us, not just hospitals and clinics. People, animals, and ecosystems are interconnected around the world, therefore there is an urgent need to use more creative approaches to understand the dynamics of rising bacterial resistance to our last–resort antibiotics.’
Professor Qiu Yang, Fujian Agriculture and Forestry University, Fuzhou, China said, ‘This research can be applied to other plasmids carrying resistance genes – it is very likely plasmids thought to have a narrow host range can in fact be spread between lots of different bacteria. Research exploring the host range of multidrug resistant plasmids is urgently needed to prevent the growing burden of drug-resistance globally.’
The study ‘Interphylum dissemination of NDM-5-positive plasmids in hospital wastewater from Fuzhou, China: a single-centre, culture-independent, plasmid transmission study’ has been published in The Lancet Microbe.
 Landmark study definitively shows that conservation actions are effective at halting and reversing biodiversity loss
Landmark study definitively shows that conservation actions are effective at halting and reversing biodiversity loss
 Researchers find oldest undisputed evidence of Earth’s magnetic field
Researchers find oldest undisputed evidence of Earth’s magnetic field
 Honorary degree recipients for 2024 announced
Honorary degree recipients for 2024 announced
 Vice-Chancellor's innovative cross-curricular programme celebrated
Vice-Chancellor's innovative cross-curricular programme celebrated
 New database sheds light on violence in Greek detention facilities
New database sheds light on violence in Greek detention facilities